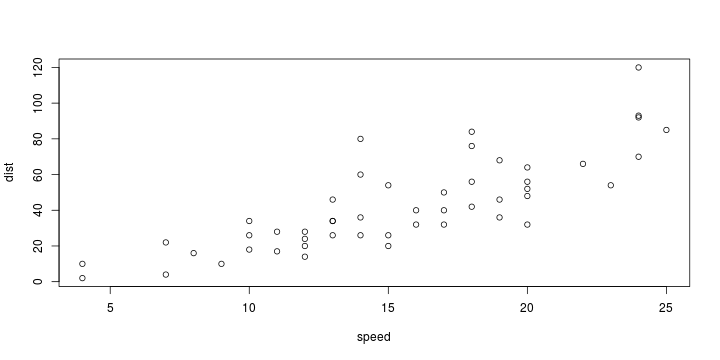
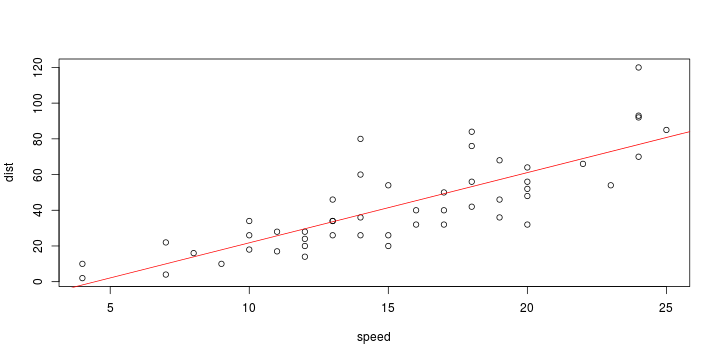
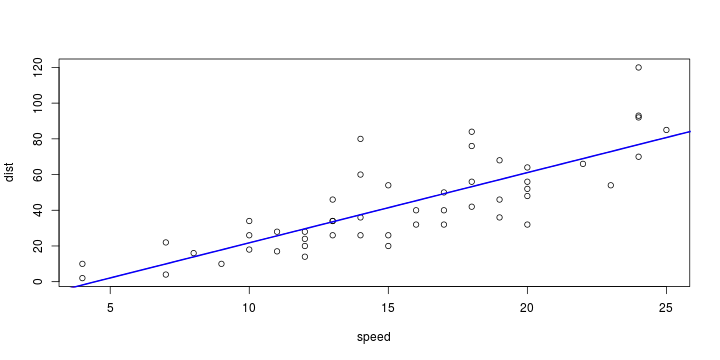
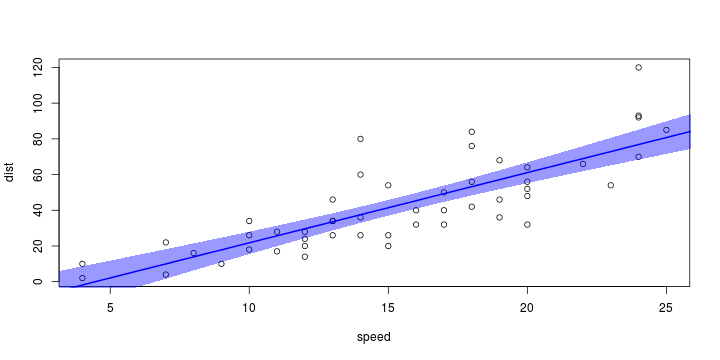
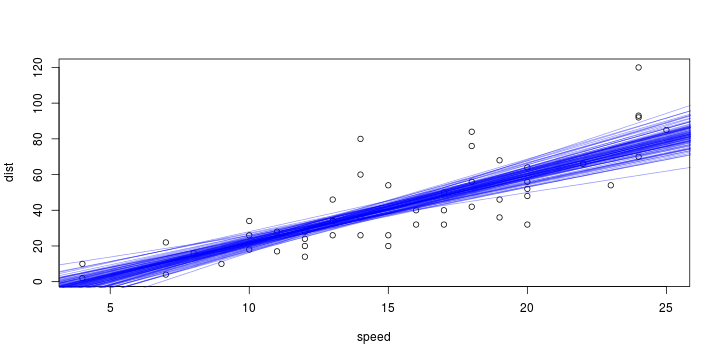
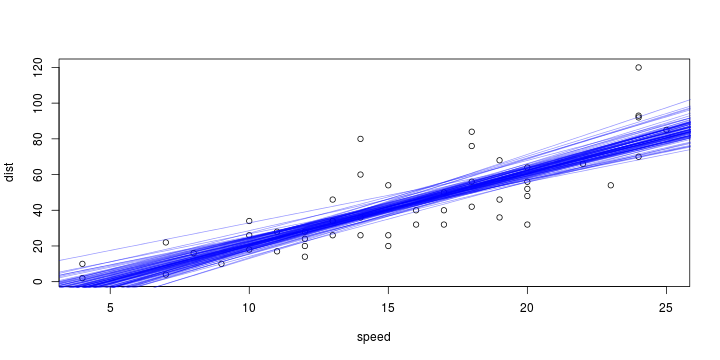
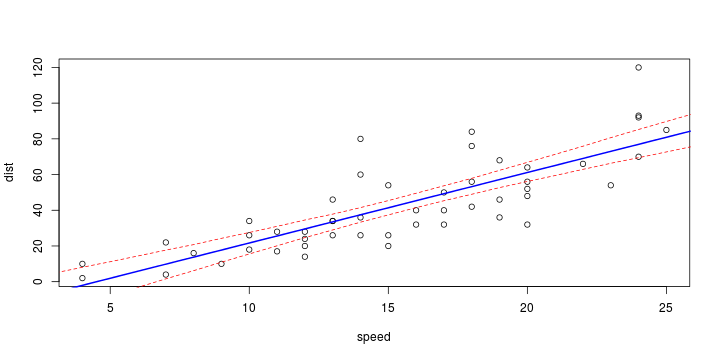
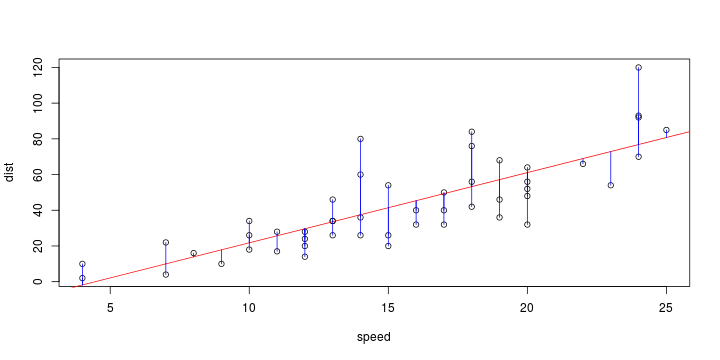
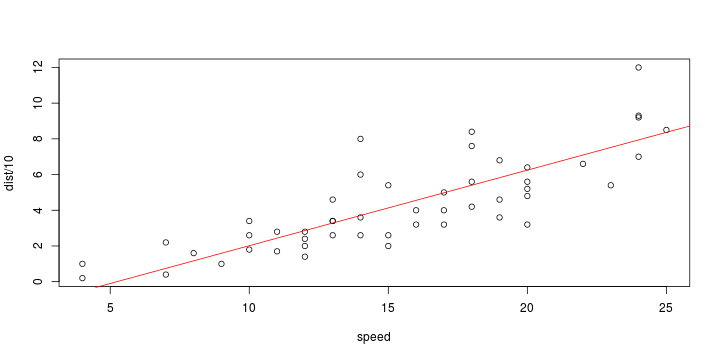
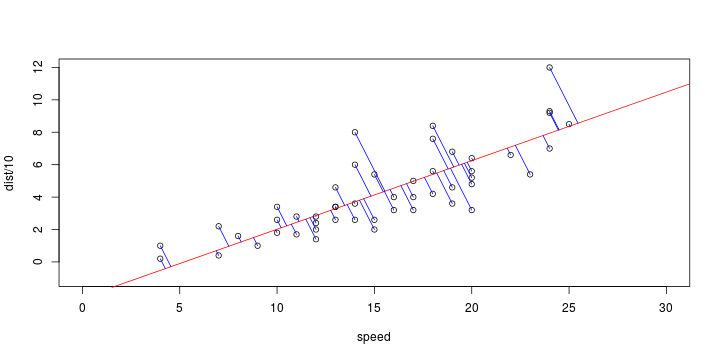
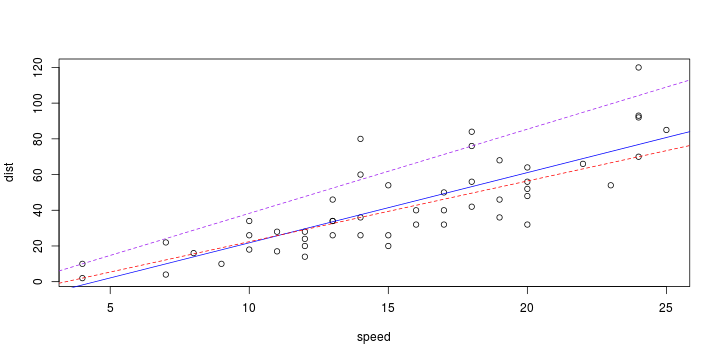
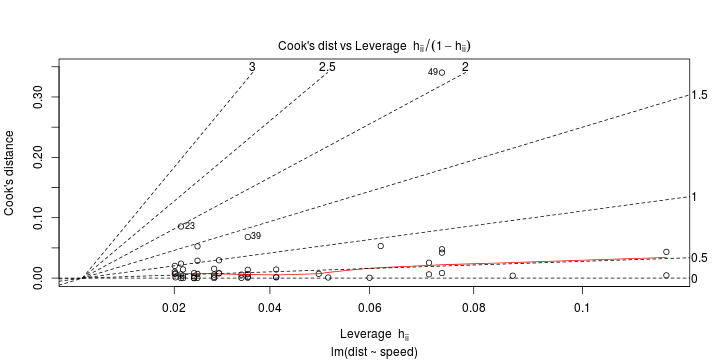
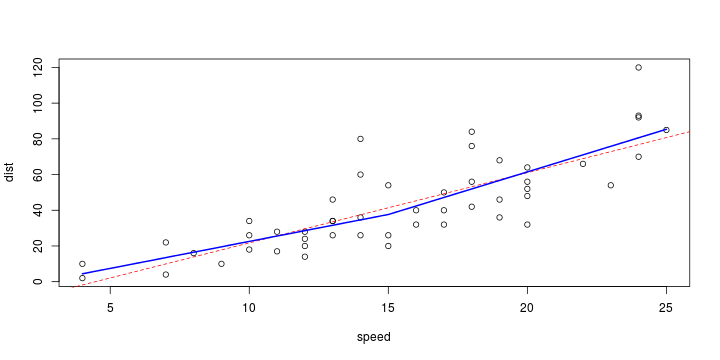
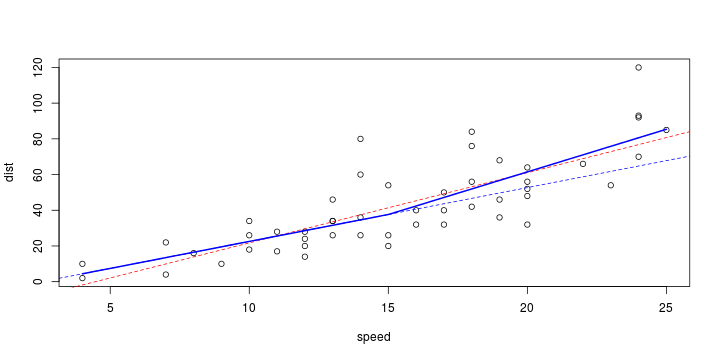
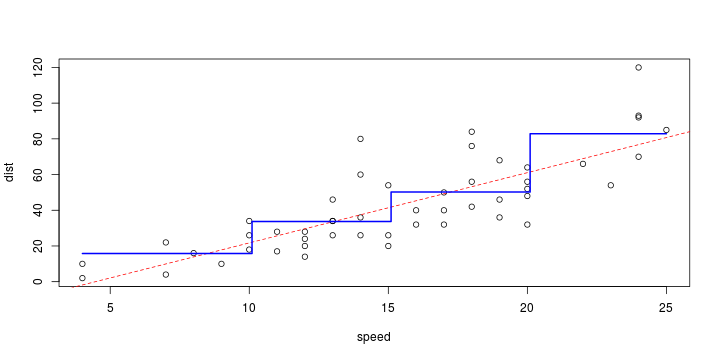
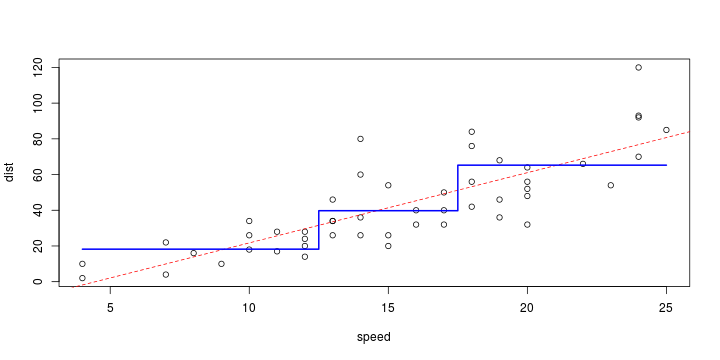
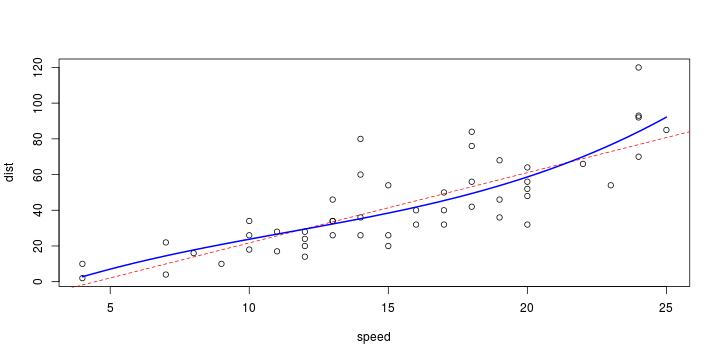
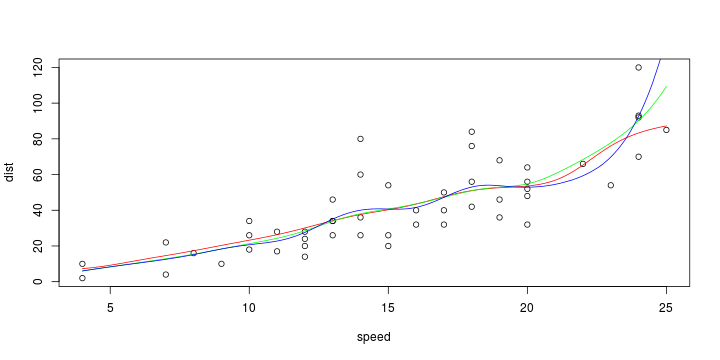
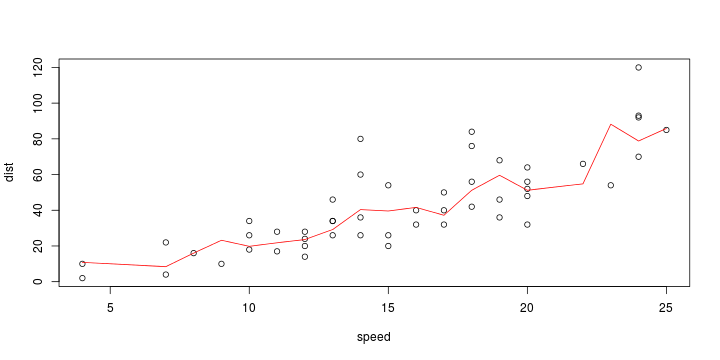
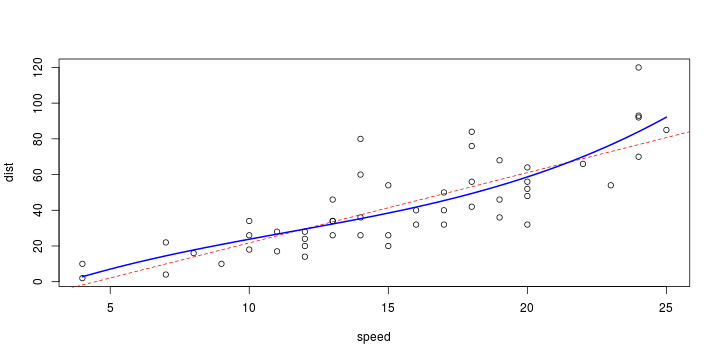
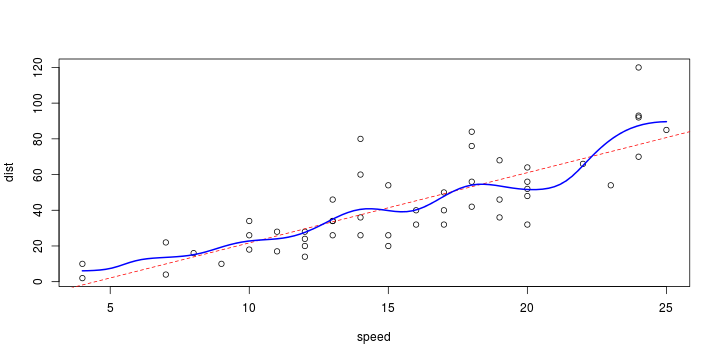
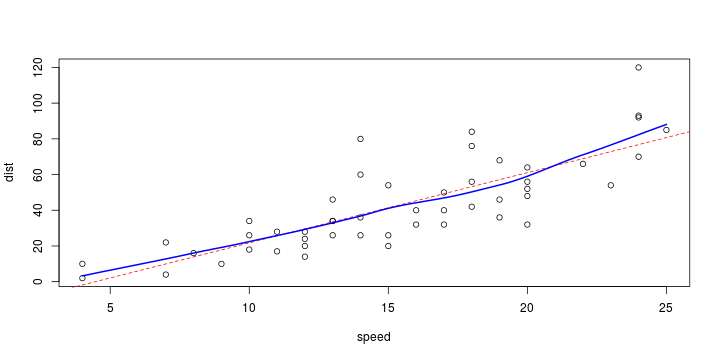
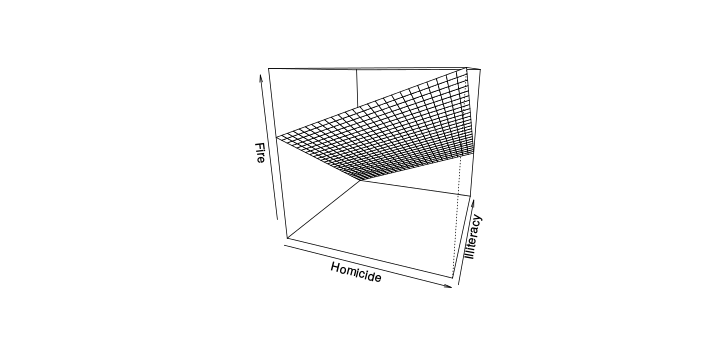
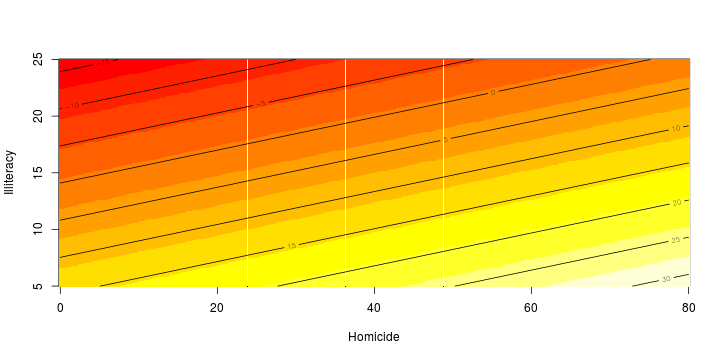
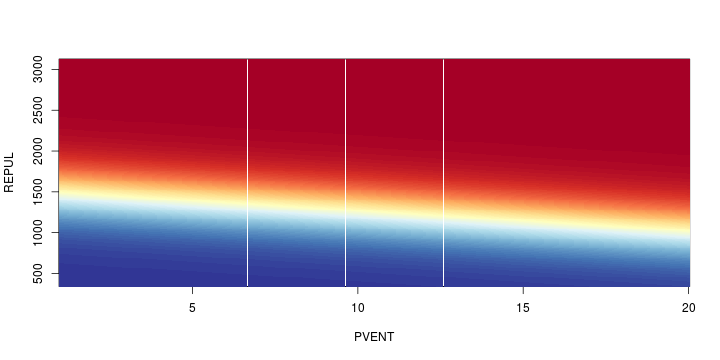
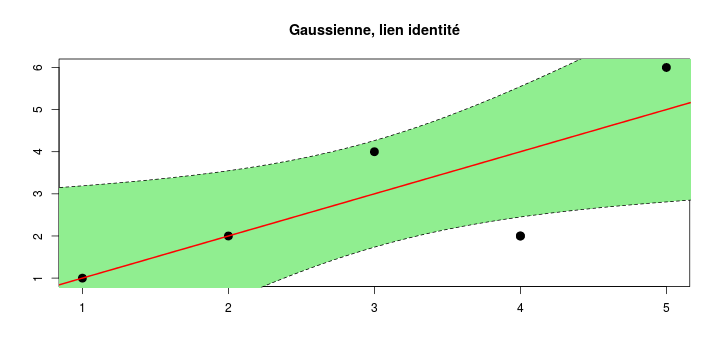
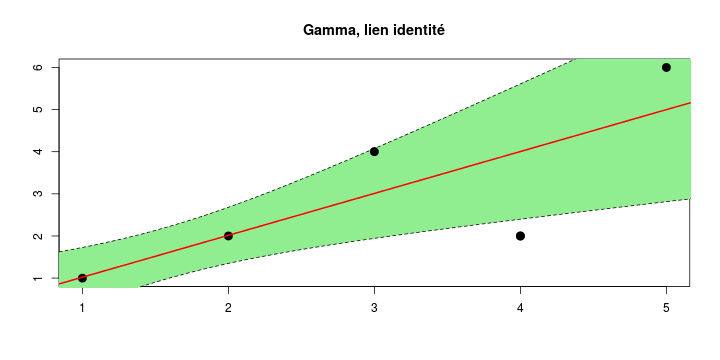
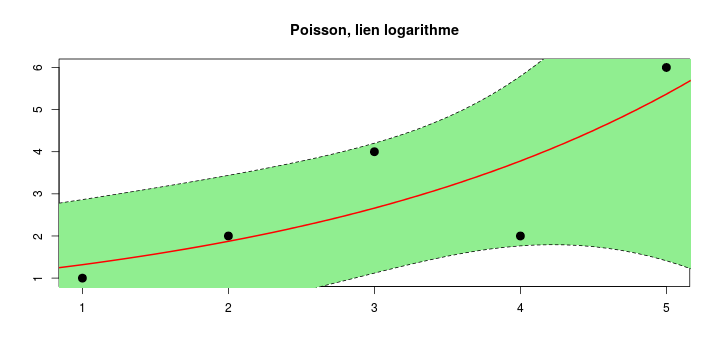
\[ y_i=\boldsymbol{x}_i^{\text{T}}\boldsymbol{\beta}+\varepsilon_i=\beta_0 + [\beta_1 x_{1,i}+\cdots+ \beta_k x_{k,i}]+\varepsilon_i \]
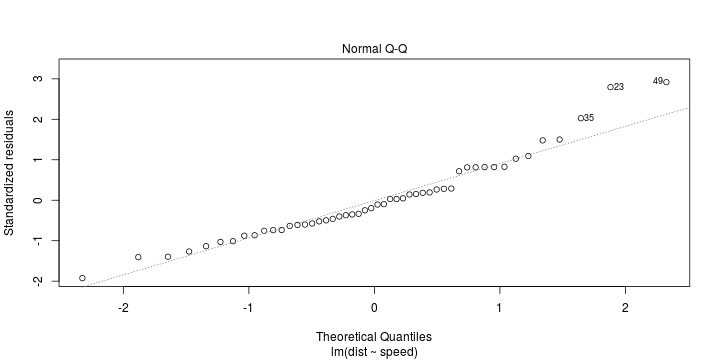
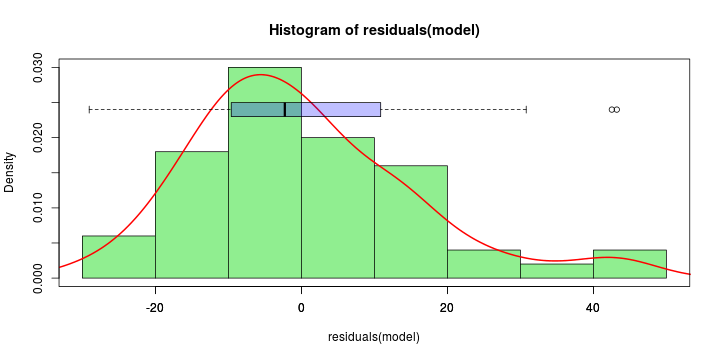
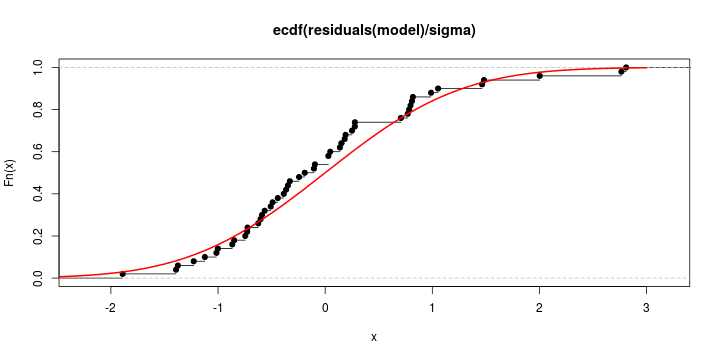
See linear model, where \(\varepsilon_i\sim\mathcal{N}(0,\sigma^2)\) i.id.
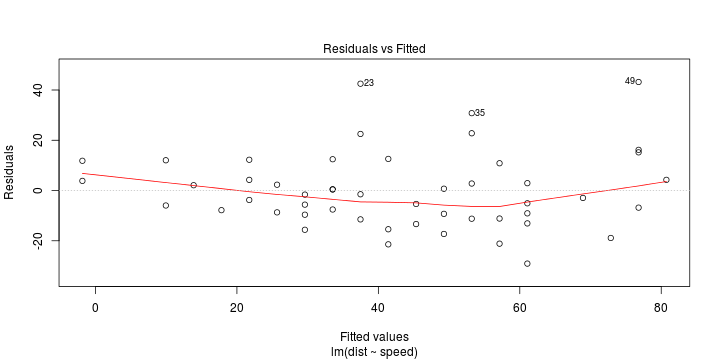
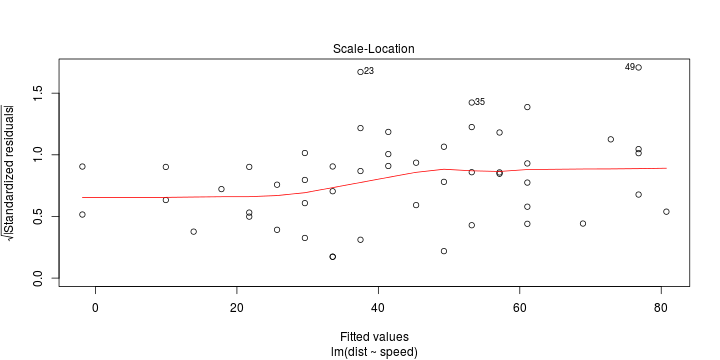
\[ (Y\vert \boldsymbol{X}=\boldsymbol{x})\sim \mathcal{N}(\boldsymbol{x}^{\text{T}}\boldsymbol{\beta}, \sigma^2) \] i.e. \[ \mathbb{E}[Y\vert\boldsymbol{X}=\boldsymbol{x}]=\boldsymbol{x}^{\text{T}}\boldsymbol{\beta} \] and homoscedastic model, \[ \text{Var}[Y\vert\boldsymbol{X}=\boldsymbol{x}]=\sigma^2 \]
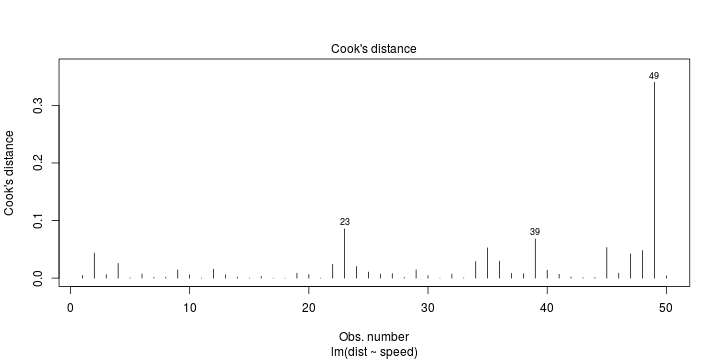
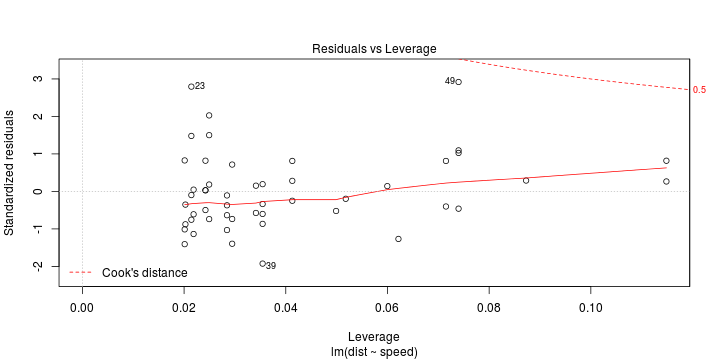
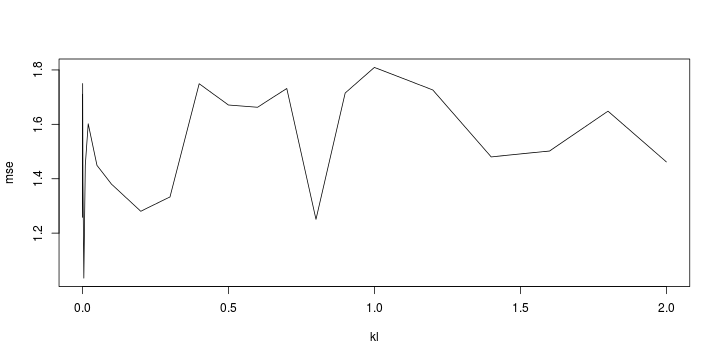
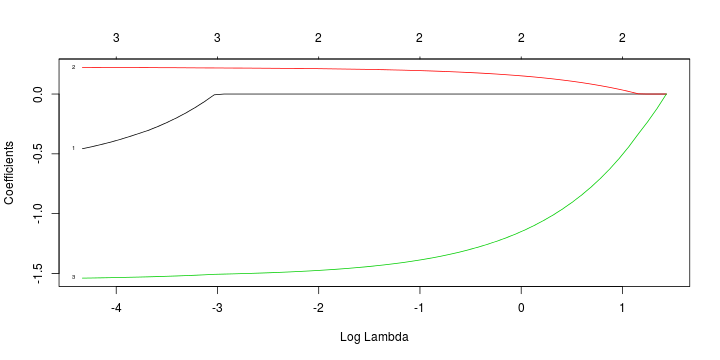
Least squares (and maximum likelihood) estimator \[ \widehat{\boldsymbol{\beta}}=\text{argmin} \left\lbrace \sum_{i=1}^n (y_i-\boldsymbol{x}_i^{\text{T}}\boldsymbol{\beta})^2 \right\rbrace =(\boldsymbol{X}^{\text{T}}\boldsymbol{X})^{-1}\boldsymbol{X}^{\text{T}}\boldsymbol{y}\]