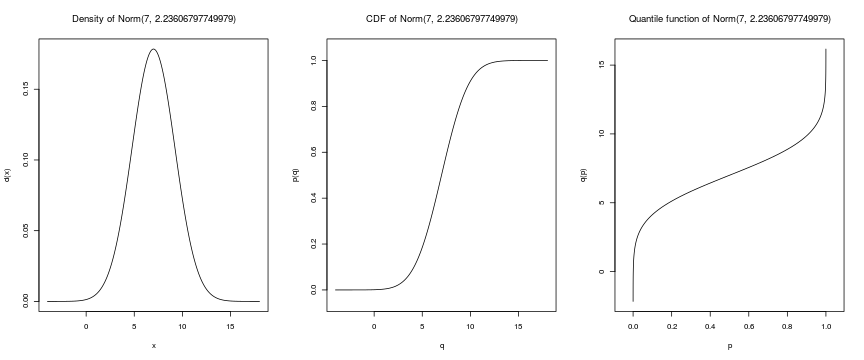
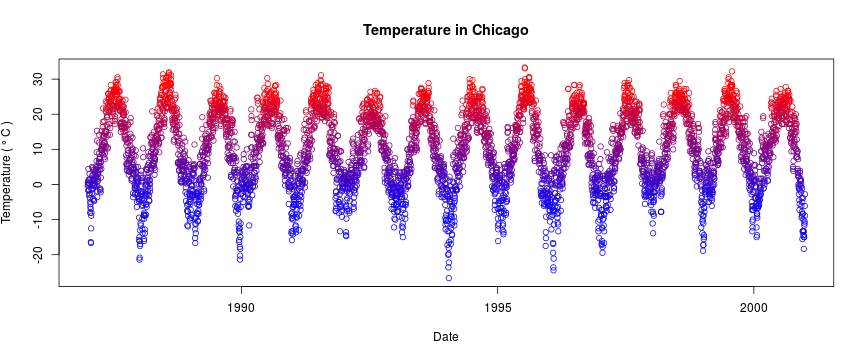
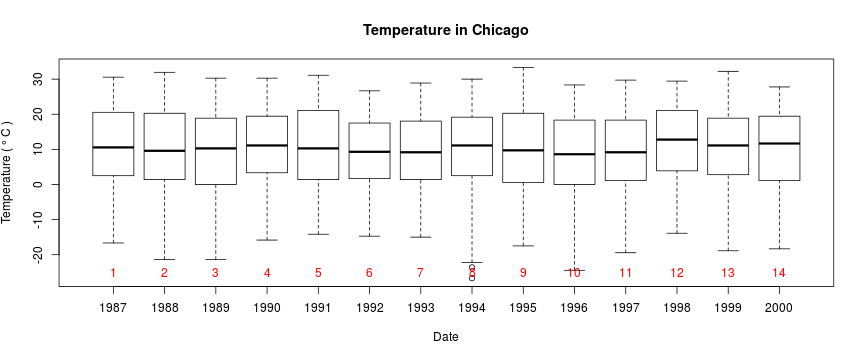
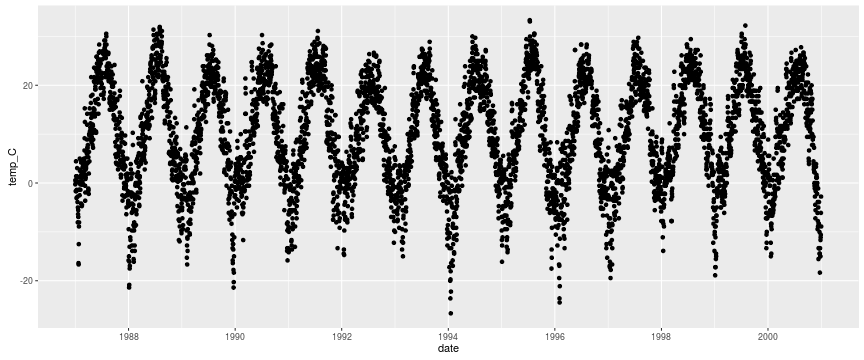
getOption("defaultPackages")
## [1] "datasets" "utils" "grDevices" "graphics" "stats" "methods"
(.packages(all.available=TRUE))
## [1] "abind" "acepack" "actuar"
## [4] "ada" "adabag" "ade4"
## [7] "ade4TkGUI" "adegraphics" "AER"
## [10] "akima" "amap" "aod"
## [13] "ape" "aplpack" "arules"
## [16] "arulesViz" "ash" "assertthat"
## [19] "base64enc" "bayesm" "bdsmatrix"
## [22] "BH" "biclust" "biglm"
## [25] "bigmemory" "bigmemory.sri" "BiocInstaller"
## [28] "bit" "bit64" "bitops"
## [31] "boot" "Boruta" "BradleyTerry2"
## [34] "brew" "brglm" "C50"
## [37] "car" "caret" "CASdatasets"
## [40] "caTools" "cba" "ChainLadder"
## [43] "chron" "circlize" "CircStats"
## [46] "classInt" "clv" "cmprsk"
## [49] "cobs" "coda" "coin"
## [52] "colorspace" "combinat" "compositions"
## [55] "corpcor" "corrplot" "cplm"
## [58] "crayon" "cshapes" "cubature"
## [61] "Cubist" "curl" "data.table"
## [64] "DBI" "deldir" "demography"
## [67] "dendroextras" "DEoptimR" "descr"
## [70] "devtools" "dichromat" "digest"
## [73] "diptest" "dismo" "distr"
## [76] "distrEx" "doBy" "doParallel"
## [79] "dplyr" "DT" "dtw"
## [82] "dynlm" "e1071" "earth"
## [85] "effects" "ellipse" "energy"
## [88] "entropy" "evaluate" "evir"
## [91] "explor" "expm" "FactoMineR"
## [94] "fastICA" "fastmatch" "fBasics"
## [97] "fda" "fGarch" "fields"
## [100] "flashClust" "flexclust" "flexmix"
## [103] "FNN" "foreach" "forecast"
## [106] "formatR" "Formula" "fpc"
## [109] "fracdiff" "FSelector" "fts"
## [112] "ftsa" "gam" "gamair"
## [115] "gamlss" "gamlss.data" "gamlss.dist"
## [118] "gbm" "gclus" "gdata"
## [121] "geosphere" "ggdendro" "ggmap"
## [124] "ggplot2" "ggthemes" "git2r"
## [127] "glmnet" "GlobalOptions" "gnm"
## [130] "goftest" "googleVis" "gplots"
## [133] "gridBase" "gridExtra" "gss"
## [136] "gstat" "gtable" "gtools"
## [139] "gWidgets" "hdrcde" "heplots"
## [142] "hexbin" "highr" "HistData"
## [145] "hmeasure" "Hmisc" "htmltools"
## [148] "htmlwidgets" "httpuv" "httr"
## [151] "igraph" "ineq" "intervals"
## [154] "ipred" "IRdisplay" "irlba"
## [157] "iterators" "its" "jpeg"
## [160] "jsonlite" "kernlab" "klaR"
## [163] "knitr" "ks" "labeling"
## [166] "LaF" "Lahman" "lars"
## [169] "latticeExtra" "lava" "lazyeval"
## [172] "leaflet" "leaps" "LearnBayes"
## [175] "lme4" "lmtest" "loa"
## [178] "locfit" "logcondens" "logicFS"
## [181] "LogicReg" "LSMonteCarlo" "lubridate"
## [184] "magrittr" "manipulate" "mapdata"
## [187] "mapproj" "maps" "maptools"
## [190] "markdown" "MASS" "MatrixModels"
## [193] "maxLik" "mboost" "mcbiopi"
## [196] "mclust" "MCMCpack" "mda"
## [199] "memoise" "mice" "microbenchmark"
## [202] "mime" "minqa" "misc3d"
## [205] "miscTools" "missMDA" "mitools"
## [208] "mix" "mlbench" "mlogit"
## [211] "mnormt" "modeltools" "mosaic"
## [214] "mosaicData" "multcomp" "multicool"
## [217] "munsell" "mvtnorm" "ncdf"
## [220] "ncdf4" "nloptr" "NLP"
## [223] "NMF" "nnet" "nnls"
## [226] "nortest" "np" "numDeriv"
## [229] "nycflights13" "odfWeave" "OpenStreetMap"
## [232] "pamr" "party" "partykit"
## [235] "pbapply" "pbkrtest" "PBSmapping"
## [238] "pcaPP" "pglm" "pixmap"
## [241] "pkgmaker" "plm" "plot3D"
## [244] "plotly" "plotmo" "plotrix"
## [247] "pls" "plyr" "pmml"
## [250] "png" "polyclip" "prabclus"
## [253] "pROC" "prodlim" "profileModel"
## [256] "proto" "proxy" "pryr"
## [259] "pscl" "psych" "psychotools"
## [262] "psychotree" "quadprog" "quantreg"
## [265] "questionr" "qvcalc" "R6"
## [268] "rainbow" "randomForest" "randtoolbox"
## [271] "ranger" "RANN" "raster"
## [274] "rasterVis" "rCharts" "RColorBrewer"
## [277] "Rcpp" "RcppArmadillo" "RcppEigen"
## [280] "RCurl" "readr" "readxl"
## [283] "registry" "relaimpo" "relimp"
## [286] "repr" "reshape" "reshape2"
## [289] "rgdal" "rgeos" "rgl"
## [292] "RgoogleMaps" "rJava" "rjson"
## [295] "RJSONIO" "rleafmap" "rmarkdown"
## [298] "RMySQL" "rngtools" "rngWELL"
## [301] "robustbase" "ROCR" "RODBC"
## [304] "roxygen2" "rpart" "rpart.plot"
## [307] "RPostgreSQL" "rPython" "RSQLite"
## [310] "rstudio" "rstudioapi" "RUnit"
## [313] "rversions" "rvest" "RWeka"
## [316] "RWekajars" "rworldmap" "rworldxtra"
## [319] "sampleSelection" "sandwich" "scales"
## [322] "scatterD3" "scatterplot3d" "sde"
## [325] "selectr" "seriation" "sfsmisc"
## [328] "shape" "shiny" "shinyapps"
## [331] "slam" "slidify" "slidifyLibraries"
## [334] "SnowballC" "sp" "spacetime"
## [337] "spam" "SparseM" "spatstat"
## [340] "spdep" "splancs" "splitstackshape"
## [343] "spls" "stabledist" "stabs"
## [346] "startupmsg" "statmod" "stringi"
## [349] "stringr" "strucchange" "subselect"
## [352] "superpc" "survey" "survival"
## [355] "SweaveListingUtils" "systemfit" "tau"
## [358] "TeachingDemos" "tensor" "tensorA"
## [361] "testthat" "textcat" "TH.data"
## [364] "tidyr" "timeDate" "timeSeries"
## [367] "tis" "tm" "trifield"
## [370] "trimcluster" "tripack" "truncreg"
## [373] "tseries" "TSP" "tweedie"
## [376] "urca" "uuid" "varSelRF"
## [379] "vcd" "vcdExtra" "verification"
## [382] "VGAM" "VGAMdata" "VIM"
## [385] "viridis" "waveslim" "weights"
## [388] "whisker" "wordcloud" "wskm"
## [391] "xlsx" "xlsxjars" "XML"
## [394] "xml2" "xtable" "xts"
## [397] "yaml" "YieldCurve" "zoo"
## [400] "base" "class" "cluster"
## [403] "codetools" "compiler" "datasets"
## [406] "foreign" "graphics" "grDevices"
## [409] "grid" "KernSmooth" "lattice"
## [412] "Matrix" "methods" "mgcv"
## [415] "nlme" "parallel" "spatial"
## [418] "splines" "stats" "stats4"
## [421] "tcltk" "tools" "utils"